
DNA Sequencing for the Community Scientist
Useful Links
-

FunDiS
Discover all of the ways to connect as a Citizen Scientist with the Fungal Diversity Survey.
-

GeneWiz
Tools for viewing Sanger Sequencing Data.
-
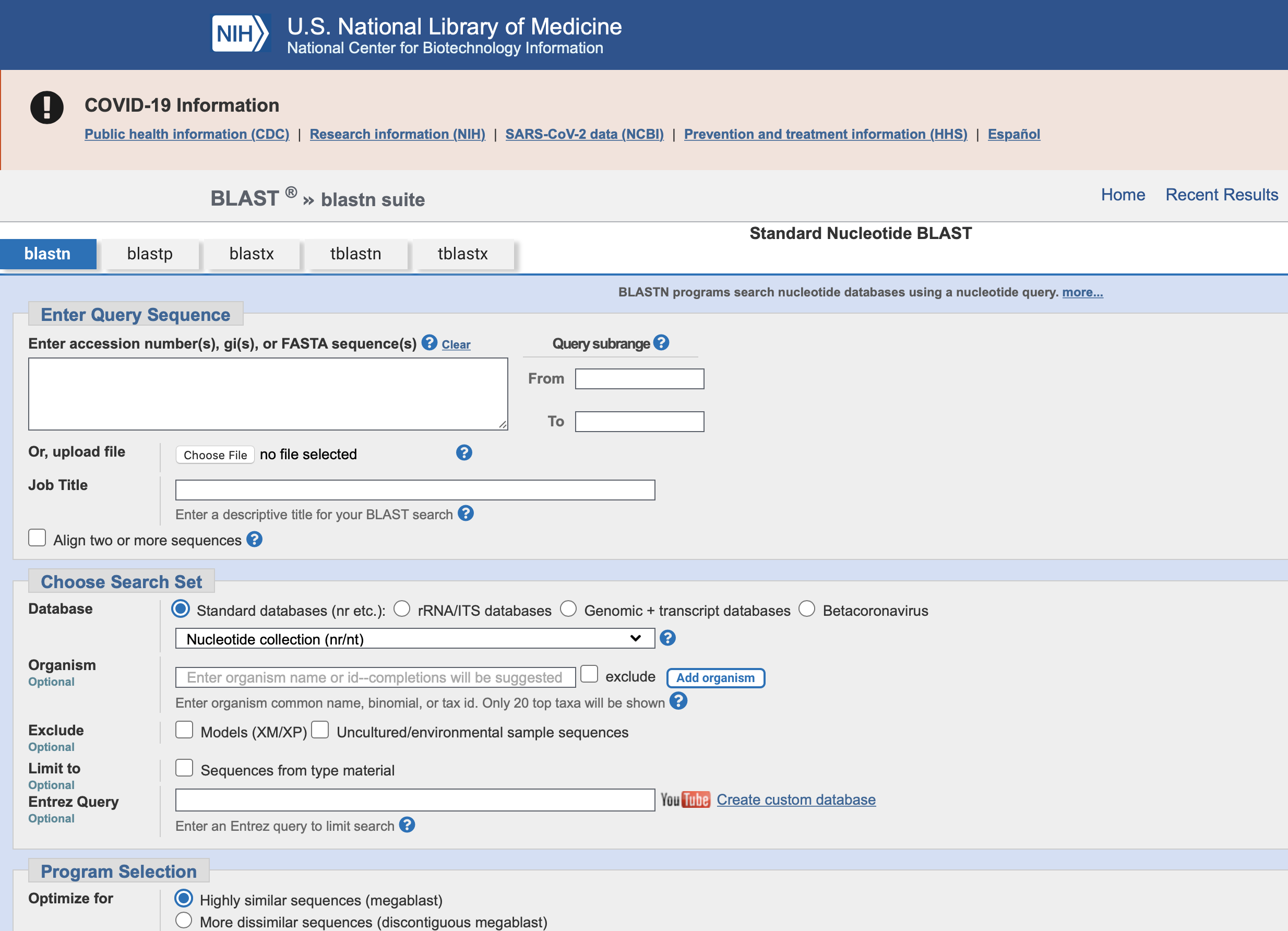
BLAST
Search engine for genetic sequences
Basic Local Alignment Search Tool
-
SnapGene Viewer
Get the FREE SnapGene Viewer to look at chromatograms or trace files from Genewiz.
-

GenBank Submission Portal
Start here to submit a named sequence to GenBank.
-

Phylogeny.fr
Create a phylogenetic tree from .FAS file.
-

MycoBank
MycoBank
Setting up a home PCR Lab
by Sigrid Jakob | PLAYLIST | SUPPLY LIST
Mushroom DNA Barcoding with Alan Rockefeller
Alan Rockefeller Contact Info:
Alan [at] mimosatherapeutics.com
Facebook.com/alan.rockefeller
Genetics 101 with Alan Rockefeller
Introduction on DNA Sequencing by Damon Tighe
Supply List
Thermal cycler
Centrifuge
Gel electrophoresis setup
2 micropipettes: 1-10 µl, 20-200 µl
Scale for very small amounts to measure out agar powder
Regular kitchen scale for weighing TBE buffer which you’ll use to make a gel block
MISC
Permanent marker
Tweezers with sharp tips
Waste container
Tube rack
Large-ish glass cup for boiling agarose and buffer
Strips of aluminum foil for mixing loading dye and fungal DNA before loading into gel
CONSUMABLES
Lots of 10µl and 200µl pipette tips, loose or in boxes (buy 1000 loose in bulk on eBay to refill boxes)
burn larger pipette tips to be used as pestles (one per sample)
Two 0.2 ml tubes per sample (loose)
Eight 0.2ml tubes in a strip - one strip for eight samples - MC Lab wants their samples in a strip, GeneWiz doesn’t
One 1.7ml tube (to pre-mix mastermix, primers and distilled water)
Two disposable non-powdered gloves (box of 100)
REAGENTS
Distilled water
0.5M NaOH - buy as crystals and dissolve - this will last you forever. To make 0.5M NaOH, measure out 5g of sodium hydroxide and dissolve it in 250ml of distilled water
Tris 8.0ph buffer - this is needs to be diluted 1:10
Mastermix (the ODIN or IBI Scientific) - keep tube upright in a small rack in a closed plastic bag in freezer
ITS1 and ITS4 primers (the ODIN, or IDT or GeneWiz especially for custom primers), store them like Mastermix. There are other primers, eg ITS4-B for basidiomycetes only (good if there might be mold contamination), but you’ll need to get them custom-mixed (GeneWiz or IDTDNA). I’ve recently been using ITS1_KYO instead of the straight ITS1 (TAGAGGAAGTAAAAGTCGTAA, custom order by GeneWiz)
Optional (tip from Damon Tighe): DMSO which can make for longer sequences
TBE buffer (miniPCR, came with kit but available elsewhere) - concentrate can stay at room temperature
Gel stain (miniPCR, came with kit, but available elsewhere) - can stay at room temperature
Loading dye (the ODIN) - keep in a small reagent rack in plastic bag in freezer
Small glass of isopropyl alcohol to sterilize hands, tweezers etc throughout (drugstore)
GenBank
Download Trace Files from GeneWiz
Use Snapgene to look at the Trace file
Verify the trace is good using BLAST
Submit to Genbank so it is available to everyone
Make phylogenetic trees to make useful data
Trace file is a chromatogram.
Primers ITS-1F (“f” stands for fungal) and ITS-4B (“4b” stands for basidiomycete).
Use SnapGene to trim and edit trace files, copy to clip board.
Go to NCBI BLAST https://blast.ncbi.nlm.nih.gov/Blast.cgi and click on Nucleotide BLAST.
Paste in data from chromatogram into query box.
Change ‘Search Set’ from Human collection to ‘Neucleotide Collection (nr/nt)’
Click BLAST to begin searching.
After data comes back scroll down and see how many listings use the same number of data points by checking the ‘Query Cover’ percentage. If using a longer primer like ITS-4B your matches may only be 80%. That is because you had more sequence data points. If the percentage reads 100% that means you have the same number of data points, or the other listing had more data points than you. Only look at sequences that are 60% or above for reliable matches.
Sort data by ‘Per. Ident’ and that shows how many bases in your sequence overlap other sequences.
Look at highest percentage with highest query cover rates to find a potential match. Finding a new species is subjective and will need further study.
Find differences and double check your chromatogram for potential problems.
Next submit the sequence with name to GenBank.
Open Note Pad and create a FASTA File.
> Mushroom name
Paste sequence…
Save File As “species.FAS'“
Open GenBank Submission Portal https://submit.ncbi.nlm.nih.gov
Eukaryotic Nuclear rRNA/ITS
Contains rRNA/ ITS region.
Required Field “Isolate”
Add name or higher classification… try and be as specific in ID. If unsure type Genus cf. species (cf. = “confer to” because no name matches better)
In Isolate field make sure to add iNat#9999999
Add field “country”
In field box type “USA: State, City, Location”
Add field “Isolation-Source”
In field box type “substrate”
Add field “Collected By”
In field box type name
Add field “Collection Date”
Add date
Add field “Primer”
Click Continue
Add Your Name
Click unpublished
Submit the sequence!
Phylogenetic Trees
LSU primer is good for comparing genera. ITS is so magnified, it is only good for comparing species.
Look for enzyme slippage in chromatograms. There will be a series of lots of the same letters then look for shadow peaks.
From BLAST Results check your top matching sequences using check boxes.
Add 15+ sequences, make sure to add one from an outgroup to give context to the tree.
Click Download, check ‘FASTA complete sequence’.
Open file with Note Pad.
Trim off long names and add location. This name will show up in the tree label.
Add in your new sequence on new line:
> Species Name
Paste in DNA sequence
Save file as a .FAS
Go to http://www.phylogeny.fr to make a Phylogenetic Tree
Click “One Click Mode”
Upload FASTA file
Click submit
Once the tree forms you can see where everything lines up. Everything on the same vertical line is the same species. The little numbers at the intersections are the likelihood of a good placement for the branches and how everything relates to each other. Anything over .7 is considered good.
Alan Rockefeller suggested this webpage for more information https://wiki.counterculturelabs.org/wiki/DNA_sequencing


